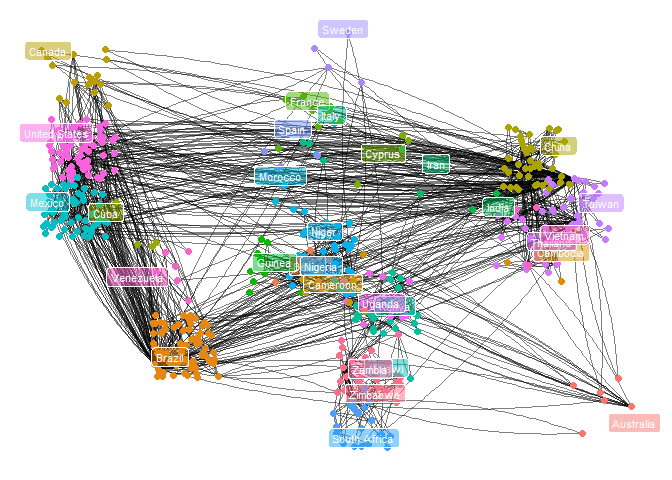
所选例子出自Modern Statistics for Modern Biology(Susan Holmes, Wolfgang Huber)
无向图的邻接矩阵是一个0-1矩阵:
library(igraph)
library(ggplot2)
library(ggnetwork)
library(network)
edges1 <- matrix(c(1,3,2,3,3,4,4,5,4,6),byrow = TRUE,ncol = 2)
### generate adjacency matrix
edges1 <- as.data.frame(edges1)
mat <- matrix(data = 0,nrow = 6,ncol = 6)
for(ii in 1:6){
mat[edges1[ii,1],edges1[ii,2]] <- 1
mat[edges1[ii,2],edges1[ii,1]] <- 1
}
### Prepare data to plot
dat_long <- reshape2::melt(mat)
dat_long$value <- as.factor(dat_long$value)
colnames(dat_long) <- c("V1","V2","value")
### plot
gg <- ggplot(dat_long)+
geom_tile(aes(V1,V2,fill=value), color="#7f7f7f")+
scale_fill_manual(values=c("white", "black"))+
coord_equal()+
labs(x=NULL, y=NULL)+
scale_x_continuous(breaks = 1:6)+
scale_y_reverse(breaks=1:6)+
theme_bw()+
theme(panel.grid=element_blank())+
theme(panel.border=element_blank())
gg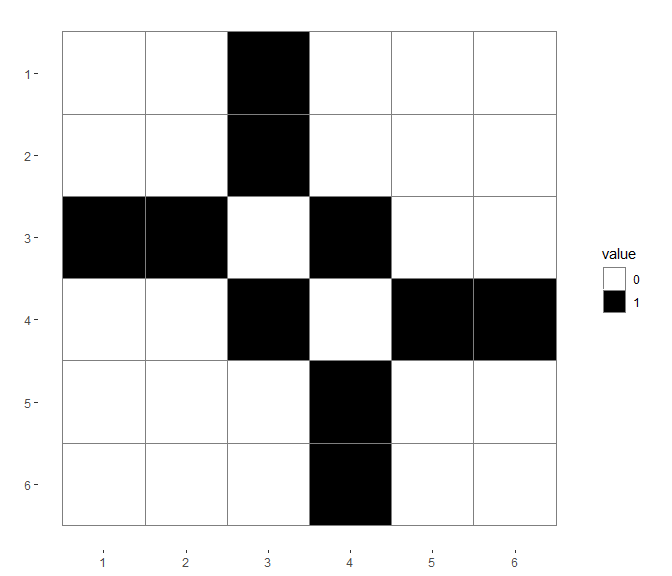
从邻接矩阵得到Graph:
g1 <- graph_from_adjacency_matrix(mat,mode = "undirected")
plot(g1,vertex.size=25,edge.width=5,vertex.color="coral")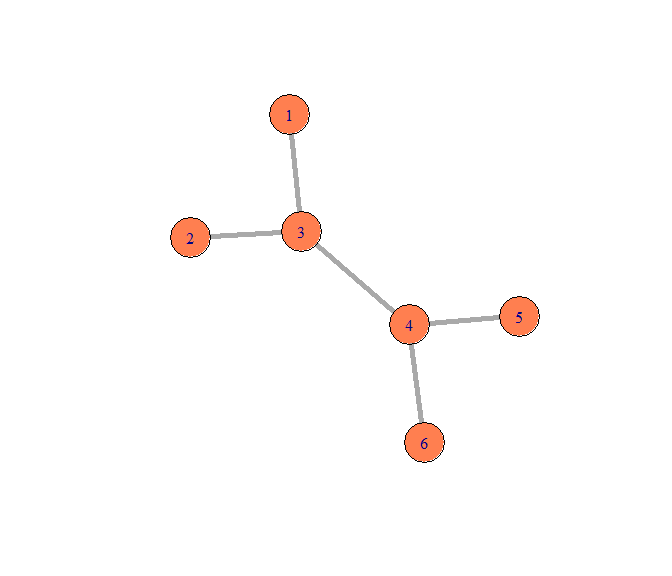
给定edgelist,得到Graph
edges1 <- matrix(c(1,3,2,3,3,4,4,5,4,6),byrow = TRUE,ncol = 2)
g1 <- graph_from_edgelist(edges1,directed = F)
plot(g1,vertex.size=25,edge.width=5,vertex.color="coral")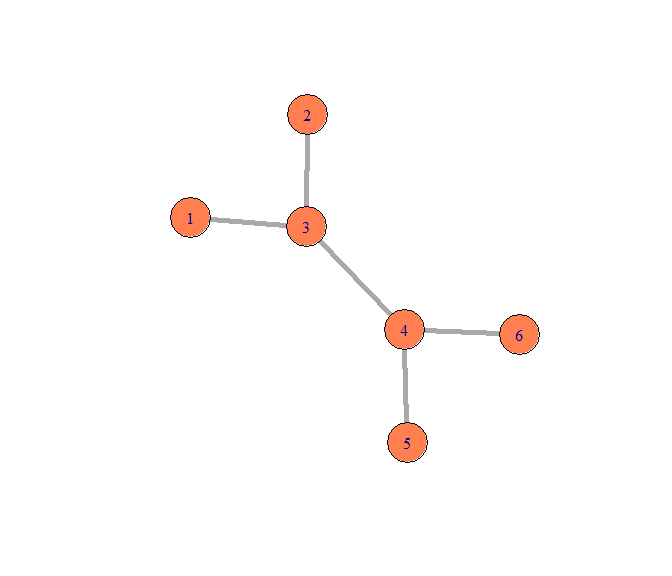
更为高级的是,从数据中计算出邻接矩阵,并且自定义可视化的layout。
library(rworldmap)
### obtain data; get the binary matrix
load("D:/tmp/Moderstatdata/data/dist2009c.RData")
country09 = attr(dist2009c, "Label")
mstree2009 = ape::mst(dist2009c)
### calculate layout from world map
mat = match(country09, countriesLow$NAME)
coords2009 = data.frame(
lat = countriesLow$LAT[mat],
lon = countriesLow$LON[mat],
country = country09)
layoutCoordinates = cbind(
x = jitter(coords2009$lon, amount = 15),
y = jitter(coords2009$lat, amount = 8))
labc = names(table(country09)[which(table(country09) > 1)])
matc = match(labc, countriesLow$NAME)
dfc = data.frame(
latc = countriesLow$LAT[matc],
lonc = countriesLow$LON[matc],
labc)
dfctrans = dfc
dfctrans[, 1] = (dfc[,1] + 31) / 93
dfctrans[, 2] = (dfc[,2] + 105) / 238
ggeo09 = ggnetwork(mstree2009, arrow.gap = 0, layout = layoutCoordinates)
###plot
ggplot(ggeo09, aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(color = "black", alpha = 0.5, curvature = 0.1) +
geom_nodes(aes(color = vertex.names), size = 2) +
theme_blank() +
geom_label(data = dfctrans, aes(x = lonc, xend = lonc, y = latc, yend = latc,
label = labc, fill = labc), colour = "white", alpha = 0.5, size = 3) +
theme(legend.position = "none")