LearnSeurat_CITE_seq
前言
CITE-seq是Rahul Satija和Peter Smibert两个组合作开发的在单细胞精度,同时测量细胞表面蛋白表达和转录组的技术。该技术原理如下:

该项技术可以用于免疫相关的单细胞测序研究中。例如, 有研究表明称:
他们在人和小鼠非小细胞肺癌中进行单细胞RNA测序,鉴定了一群DC,并将其命名为“富含免疫调节分子的成熟DC”(mregDC),这是由于它们共表达了免疫调节基因(Cd274,Pdcd1lg2和Cd200)和成熟基因(Cd40,Ccr7和Il12b)。
这段中文报道来自小柯机器人
Rahul Satija组开发的软件Seurat有一个教程,可以分析CITE-seq数据。本文基于该教程对该类型数据的分析进行说明。
数据载入
首先我们需要获取数据,该数据集取样为8617个脐带血单核细胞,包含了表达谱数据和11个抗体来源标签数据(antibody-derived tags ,ADT)。
library(Seurat)
library(SeuratData)
library(ggplot2)
library(patchwork)
### AvailableData() check avaliable data: we choose cbmc
### InstallData('cbmc')
library(cbmc.SeuratData)
data("cbmc")
### expression matrix
cbmc[["RNA"]]@counts[1:10,1:10]## 10 x 10 sparse Matrix of class "dgCMatrix"
##
## A1BG . . . . . . . . . .
## A1BG-AS1 . . . . . . . . . .
## A1CF . . . . . . . . . .
## A2M . . . . . . . . . .
## A2M-AS1 . . . . . . . 1 . .
## A2ML1 . . . . . . . . . .
## A4GALT . . . . . . . . . .
## A4GNT . . . . . . . . . .
## AAAS . . . . . . . . . 1
## AACS . . . . . . . . . .## 10 x 10 sparse Matrix of class "dgCMatrix"
##
## CD3 60 52 89 55 63 82 53 42 103 56
## CD4 72 49 112 66 80 78 63 59 122 70
## CD8 76 59 61 56 94 57 61 55 64 80
## CD45RA 575 3943 682 378 644 479 487 472 540 535
## CD56 64 68 87 58 104 44 64 48 136 91
## CD16 161 107 117 82 168 92 77 99 235 131
## CD11c 77 65 65 44 92 63 70 75 106 69
## CD14 206 129 169 136 164 122 112 111 206 204
## CD19 70 665 79 49 81 44 60 58 61 107
## CD34 179 79 78 83 152 103 79 86 144 193## [1] "RNA"根据基因表达进行聚类
注意在默认参数的情况下,下述操作时对Default Assay进行的
# standard log-normalization
cbmc <- NormalizeData(cbmc)
# choose ~1k variable features
cbmc <- FindVariableFeatures(cbmc)
# standard scaling (no regression)
cbmc <- ScaleData(cbmc)
# Run PCA, select 13 PCs for tSNE visualization and graph-based clustering
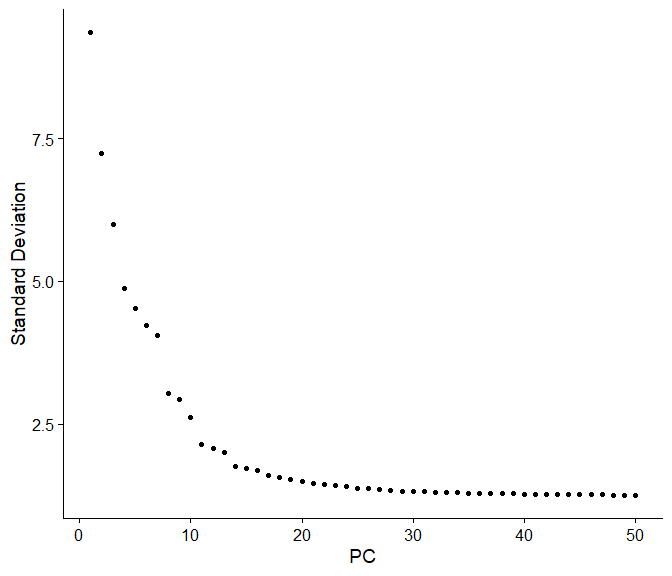
cbmc <- RunPCA(cbmc, verbose = FALSE)下面的图是根据标准差来选择PCs
聚类和t-SNE降维
## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 8617
## Number of edges: 347548
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8592
## Number of communities: 19
## Elapsed time: 3 secondscbmc <- RunTSNE(cbmc, dims = 1:25, method = "FIt-SNE")
# Find the markers that define each cluster, and use these to annotate the clusters, we use
# max.cells.per.ident to speed up the process
cbmc.rna.markers <- FindAllMarkers(cbmc, max.cells.per.ident = 100, min.diff.pct = 0.3, only.pos = TRUE)
# Note, for simplicity we are merging two CD14+ Monocyte clusters (that differ in expression of
# HLA-DR genes) and NK clusters (that differ in cell cycle stage)
new.cluster.ids <- c("Memory CD4 T", "CD14+ Mono", "Naive CD4 T", "NK", "CD14+ Mono", "Mouse", "B",
"CD8 T", "CD16+ Mono", "T/Mono doublets", "NK", "CD34+", "Multiplets", "Mouse", "Eryth", "Mk",
"Mouse", "DC", "pDCs")
names(new.cluster.ids) <- levels(cbmc)
cbmc <- RenameIdents(cbmc, new.cluster.ids)我们看看聚类结果:
## Warning: Using `as.character()` on a quosure is deprecated as of rlang 0.3.0.
## Please use `as_label()` or `as_name()` instead.
## This warning is displayed once per session.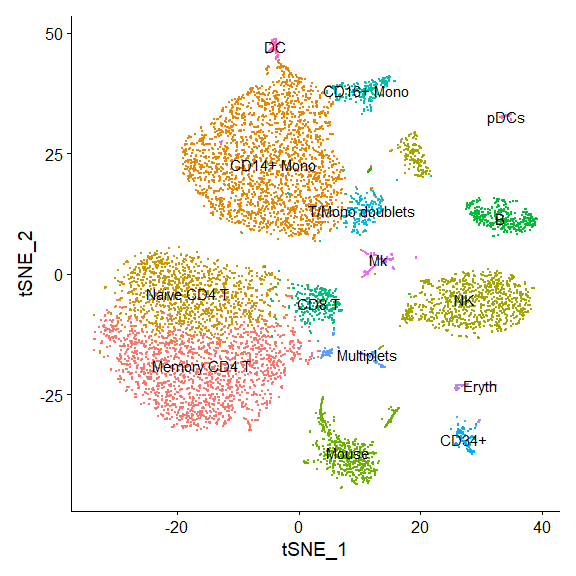
蛋白表达数据处理
Seurat3的assay实现多个组学或者模态的数据的存储和获取。 代码里的注释来自Seurat官网。
# Now we can repeat the preprocessing (normalization and scaling) steps that we typically run
# with RNA, but modifying the 'assay' argument. For CITE-seq data, we do not recommend typical
# LogNormalization. Instead, we use a centered log-ratio (CLR) normalization, computed
# independently for each feature. This is a slightly improved procedure from the original
# publication, and we will release more advanced versions of CITE-seq normalizations soon.
cbmc <- NormalizeData(cbmc, assay = "ADT", normalization.method = "CLR")
cbmc <- ScaleData(cbmc, assay = "ADT")在RNA表达谱的降维Embedding中同时展示展示蛋白表达水平和基因表达水平:
散点图:横纵轴为降维的坐标:
# in this plot, protein (ADT) levels are on top, and RNA levels are on the bottom
FeaturePlot(cbmc,
features = c("adt_CD3", "adt_CD11c",
"adt_CD8", "adt_CD16",
"CD3E", "ITGAX", "CD8A", "FCGR3A"),
min.cutoff = "q05",
max.cutoff = "q95",
ncol = 2)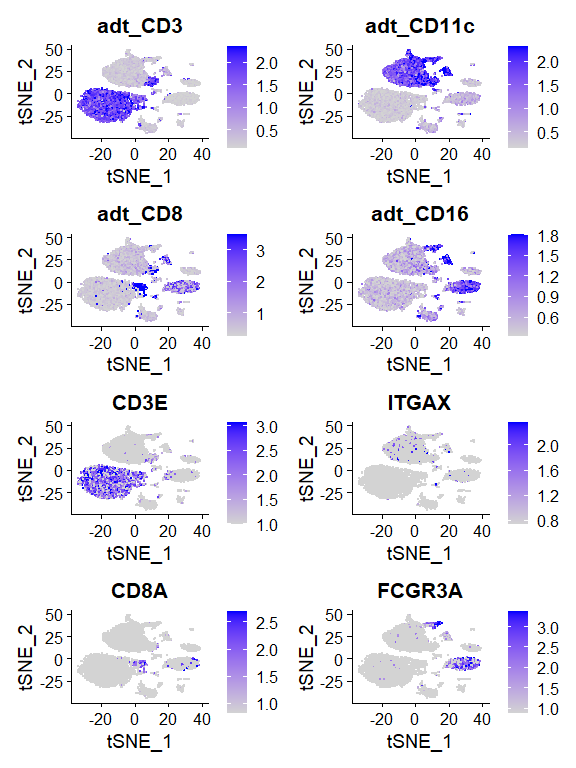
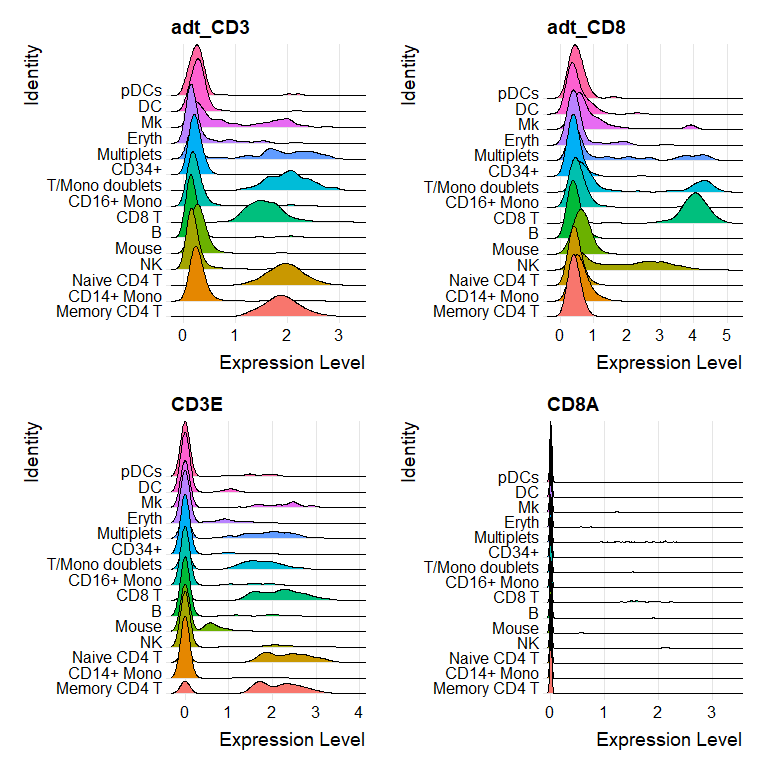
Ridge Plot:
散点图:横纵轴为表达量;这个类似于FACS
# Draw ADT scatter plots (like biaxial plots for FACS). Note that you can even 'gate' cells if
# desired by using HoverLocator and FeatureLocator
FeatureScatter(cbmc, feature1 = "adt_CD19", feature2 = "adt_CD3")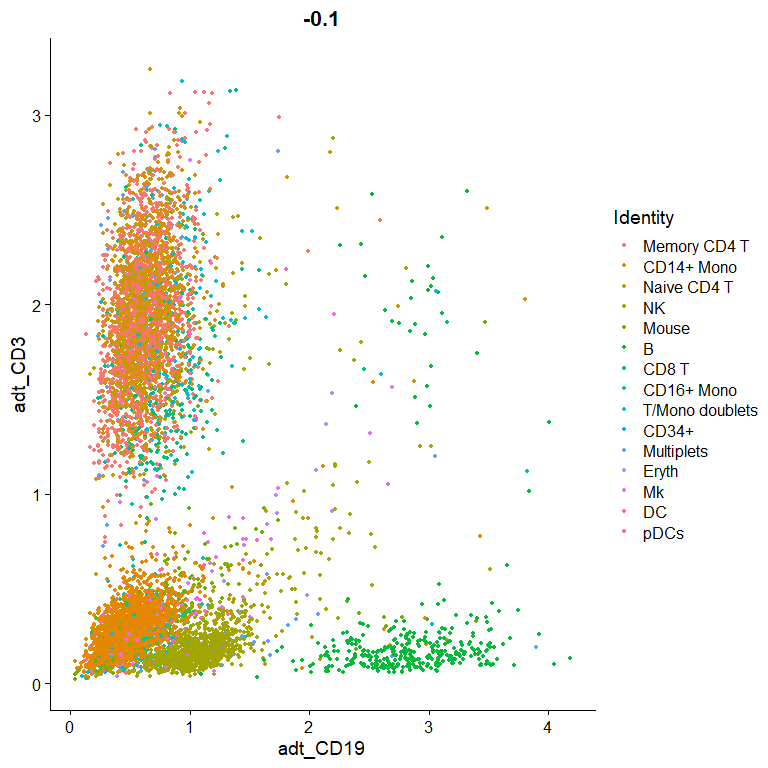
我们也可以看看蛋白表达和基因表达的关系:
# view relationship between protein and RNA
FeatureScatter(cbmc, feature1 = "adt_CD3", feature2 = "CD3E")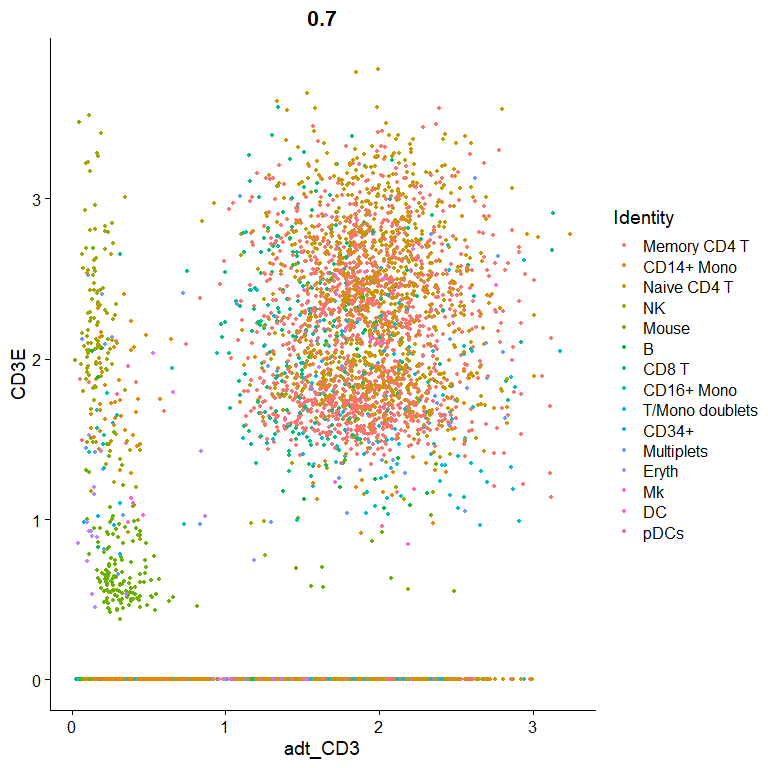
我们可以看看T细胞:
# Let's plot CD4 vs CD8 levels in T cells
tcells <- subset(cbmc, idents = c("Naive CD4 T", "Memory CD4 T", "CD8 T"))
FeatureScatter(tcells, feature1 = "adt_CD4", feature2 = "adt_CD8")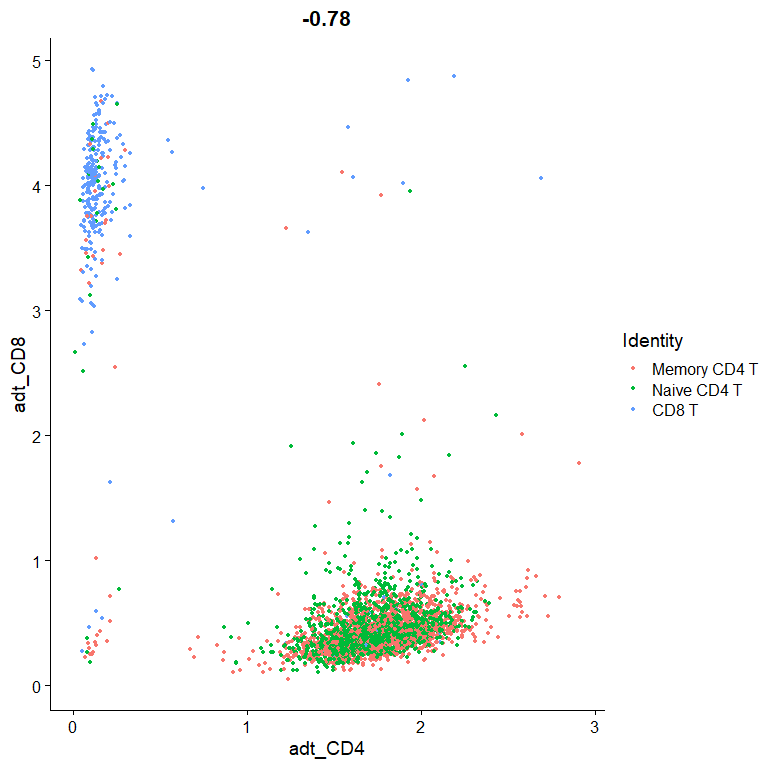
选没有标准化的原始数据我们看看,坐标轴的间距太大,会有misleading
# # Let's look at the raw (non-normalized) ADT counts. You can see the values are quite high,
# particularly in comparison to RNA values. This is due to the significantly higher protein copy
# number in cells, which significantly reduces 'drop-out' in ADT data
FeatureScatter(tcells, feature1 = "adt_CD4", feature2 = "adt_CD8", slot = "counts")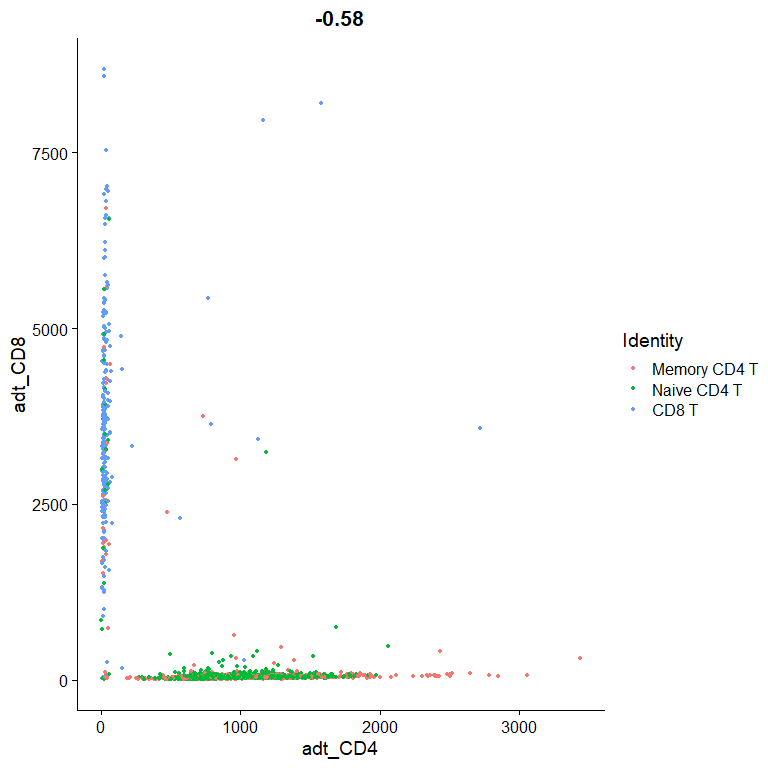
这里还是可以观察到dropouts现象的,据原作者说: > If you look a bit more closely, you’ll see that our CD8 T cell cluster is enriched for CD8 T cells, but still contains many CD4+ CD8- T cells. This is because Naive CD4 and CD8 T cells are quite similar transcriptomically, and the RNA dropout levels for CD4 and CD8 are quite high. This demonstrates the challenge of defining subtle immune cell differences from scRNA-seq data alone.
画热图,Seurat3 加了 downsample的功能。
# Downsample the clusters to a maximum of 300 cells each (makes the heatmap easier to see for small clusters)
cbmc.small <- subset(cbmc, downsample = 300)
# Find protein markers for all clusters, and draw a heatmap
adt.markers <- rownames(cbmc.small[["ADT"]]@counts)我们可以看看Seurat热图的默认配色(三个冒号可以看更为底层的函数), 个人觉得并不好看。
# using code from RColorBrewer to demo the palette
n = 200
par(mfrow=c(3,1))
image(
1:n, 1, as.matrix(1:n),
col = Seurat:::PurpleAndYellow(k=n),
xlab = "PurpleAndYellow n", ylab = "", xaxt = "n", yaxt = "n", bty = "n"
)
image(
1:n, 1, as.matrix(1:n),
col = colorRampPalette(c("navy", "white", "firebrick3"))(n),
xlab = "NavyWhite3Firebrick3 n", ylab = "", xaxt = "n", yaxt = "n", bty = "n"
)
image(
1:n, 1, as.matrix(1:n),
col = colorRampPalette(RColorBrewer::brewer.pal(11,"RdBu"))(n),
xlab = "RdBu n", ylab = "", xaxt = "n", yaxt = "n", bty = "n"
)
把默认配色换掉,见
mypal <- rev(colorRampPalette(RColorBrewer::brewer.pal(11,"RdBu"))(256))
#mypal2 <- colorRampPalette(c("navy", "white", "firebrick3"))(256)
DoHeatmap(cbmc.small,
features = unique(adt.markers),
assay = "ADT",
angle = 90,size = 3)+
scale_fill_gradientn(colors = mypal)## Scale for 'fill' is already present. Adding another scale for 'fill', which
## will replace the existing scale.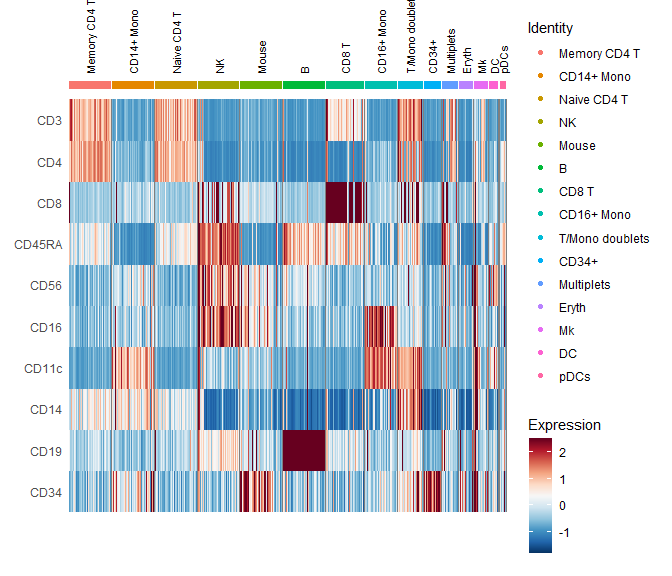
去除细胞杂质,
# You can see that our unknown cells co-express both myeloid and lymphoid markers (true at the
# RNA level as well). They are likely cell clumps (multiplets) that should be discarded. We'll
# remove the mouse cells now as well
cbmc <- subset(cbmc, idents = c("Multiplets", "Mouse"), invert = TRUE)直接根据蛋白质表达水平进行聚类
# Because we're going to be working with the ADT data extensively, we're going to switch the
# default assay to the 'CITE' assay. This will cause all functions to use ADT data by default,
# rather than requiring us to specify it each time
DefaultAssay(cbmc) <- "ADT"
cbmc <- RunPCA(cbmc, features = rownames(cbmc), reduction.name = "pca_adt", reduction.key = "pca_adt_",
verbose = FALSE)再来看PCA(其实这里算是degenrate到线性组合了)
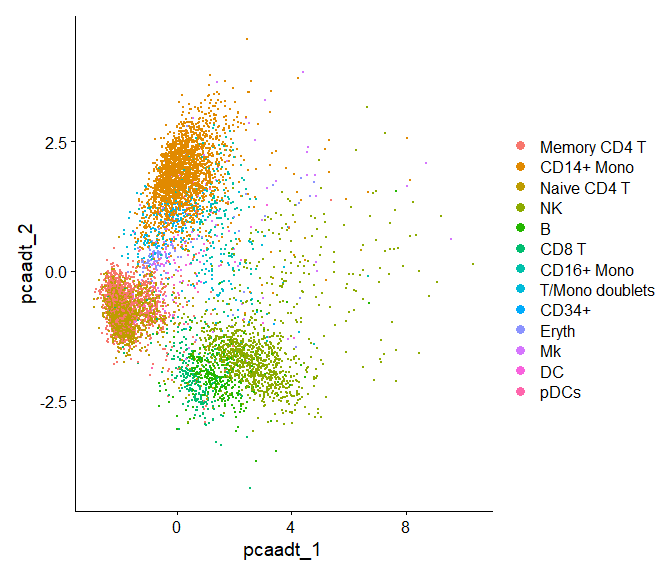
# Since we only have 10 markers, instead of doing PCA, we'll just use a standard euclidean
# distance matrix here. Also, this provides a good opportunity to demonstrate how to do
# visualization and clustering using a custom distance matrix in Seurat
adt.data <- GetAssayData(cbmc, slot = "data")
adt.dist <- dist(t(adt.data))
# Before we recluster the data on ADT levels, we'll stash the RNA cluster IDs for later
cbmc[["rnaClusterID"]] <- Idents(cbmc)
# Now, we rerun tSNE using our distance matrix defined only on ADT (protein) levels.
cbmc[["tsne_adt"]] <- RunTSNE(adt.dist, assay = "ADT", reduction.key = "adtTSNE_")
cbmc[["adt_snn"]] <- FindNeighbors(adt.dist)$snn
cbmc <- FindClusters(cbmc, resolution = 0.2, graph.name = "adt_snn")## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 7895
## Number of edges: 258146
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.9491
## Number of communities: 11
## Elapsed time: 2 seconds# We can compare the RNA and protein clustering, and use this to annotate the protein clustering
# (we could also of course use FindMarkers)
clustering.table <- table(Idents(cbmc), cbmc$rnaClusterID)
clustering.table##
## Memory CD4 T CD14+ Mono Naive CD4 T NK B CD8 T CD16+ Mono
## 0 1754 0 1217 29 0 27 0
## 1 0 2189 0 4 0 0 30
## 2 3 0 2 890 3 1 0
## 3 0 4 0 2 319 0 2
## 4 24 0 18 4 1 243 0
## 5 1 27 4 157 2 2 10
## 6 4 5 0 1 0 0 0
## 7 4 59 4 0 0 0 9
## 8 0 9 0 2 0 0 179
## 9 0 0 1 0 0 0 0
## 10 1 0 2 0 25 0 0
##
## T/Mono doublets CD34+ Eryth Mk DC pDCs
## 0 5 2 4 24 1 2
## 1 1 1 5 25 55 0
## 2 0 1 3 7 2 1
## 3 0 2 2 3 0 0
## 4 0 0 1 2 0 0
## 5 56 0 9 16 6 2
## 6 1 113 81 16 5 0
## 7 117 0 0 2 0 1
## 8 0 0 0 1 0 0
## 9 0 0 0 0 1 43
## 10 2 0 0 0 0 0下面这个embeding 还是根据ADT来的(不过只要marker连续,只有10个也没有关系?)
new.cluster.ids <- c("CD4 T", "CD14+ Mono", "NK", "B", "CD8 T", "NK", "CD34+", "T/Mono doublets",
"CD16+ Mono", "pDCs", "B")
names(new.cluster.ids) <- levels(cbmc)
cbmc <- RenameIdents(cbmc, new.cluster.ids)
tsne_rnaClusters <- DimPlot(cbmc, reduction = "tsne_adt", group.by = "rnaClusterID") + NoLegend()
tsne_rnaClusters <- tsne_rnaClusters + ggtitle("Clustering based on scRNA-seq") + theme(plot.title = element_text(hjust = 0.5))
tsne_rnaClusters <- LabelClusters(plot = tsne_rnaClusters, id = "rnaClusterID", size = 4)
tsne_adtClusters <- DimPlot(cbmc, reduction = "tsne_adt", pt.size = 0.5) + NoLegend()
tsne_adtClusters <- tsne_adtClusters + ggtitle("Clustering based on ADT signal") + theme(plot.title = element_text(hjust = 0.5))
tsne_adtClusters <- LabelClusters(plot = tsne_adtClusters, id = "ident", size = 4)
# Note: for this comparison, both the RNA and protein clustering are visualized on a tSNE
# generated using the ADT distance matrix.
wrap_plots(list(tsne_rnaClusters, tsne_adtClusters), ncol = 2)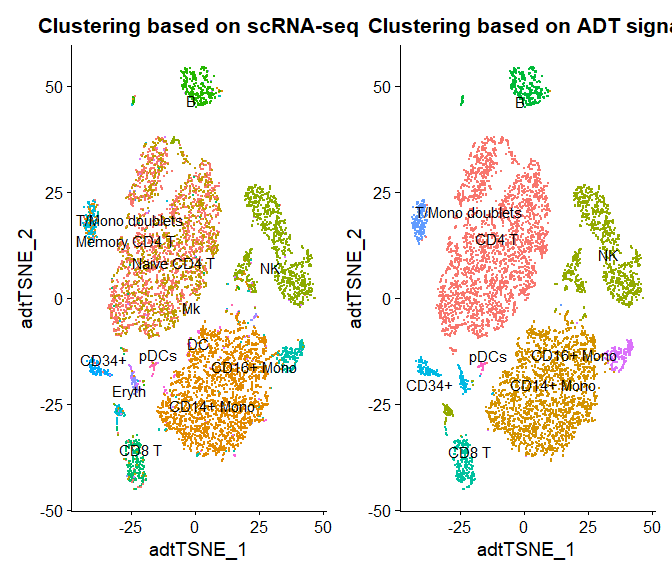 对于该结果,作者是这么解释的:
对于该结果,作者是这么解释的:
The ADT-based clustering yields similar results, but with a few differences + Clustering is improved for CD4/CD8 T cell populations, based on the robust ADT data for + CD4, CD8, CD14, and CD45RA + However, some clusters for which the ADT data does not contain good distinguishing protein markers (i.e. Mk/Ery/DC) lose separation You can verify this using FindMarkers at the RNA level, as well
更多
pbmc 10k的细胞也提供了CITE-seq的多模态数据,具体细节,请看Seurat官方教程。